INTRODUCTION
The regulation of protein synthesis is an important part of the regulation of gene expression. Regulation of mRNA translation controls the levels of particular proteins that are synthesized upon demand, such as synthesis of the different chains of globin in hemoglobin, or the production of insulin from stored insulin mRNAs in response to blood glucose levels, to name a few. The control of the cell cycle and cell proliferation also involves regulation of protein synthesis, and malignant transformation of cells involves loss of certain translational regulatory controls. In fact, several translation initiation factors are over-expressed in certain cancers and play key roles in tumor development and progression. The process of protein synthesis and important examples of its regulation are now understood at the molecular level. We will discuss the mechanism and regulation of protein synthesis, elucidating this complex area of gene regulation with specific examples.
Many viruses compete with their infected host cell and often dominate the protein synthetic machinery to maintain viral production and thwart innate (intracellular) anti-viral responses. For many viruses, the inhibition of host cell protein synthesis is an important component of their ability to propagate and destroy the infected cell. The infected cell, in turn, responds by enacting antiviral activities that include the production of potent biological molecules such as β-interferon that function, in part, to inhibit protein synthesis. Finally, a large proportion of antibiotics currently in use or under development inhibit protein synthesis in bacteria but not animal cells by exploiting differences in the structure of prokaryotic and eukaryotic ribosomes.
GENETIC CODE
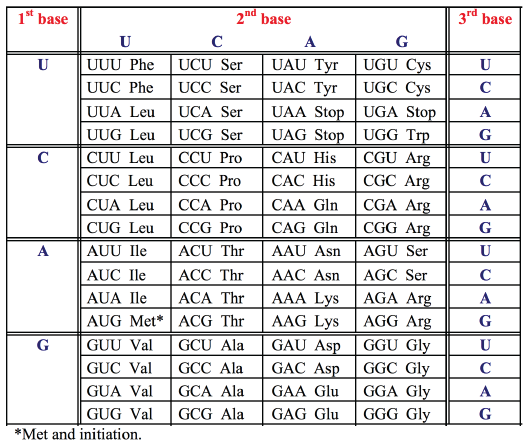
Since the genetic code is read in triplets (codons) comprising three of the four bases, there are 43 or 64 possible triplets encoding the 20 amino acids. All but 3 of these 64 codons specify amino acids. Since there are 61 codons specifying only 20 amino acids, the same amino acid may be encoded by more than one codon. The genetic code is therefore degenerate. The code is read by transfer RNAs (tRNAs), which are adapter molecules that decode the base sequence of an mRNA into the amino acid sequence of a protein. For each amino acid there is at least one corresponding tRNA which transports that amino acid to the ribosome and recognizes the particular codon(s) in the mRNA.- Each amino acid is specified by a group of three nucleotides (codon)
- The genetic code is redundant (degenerate) - most amino acids have more than one codon (only methionine "start" AUG, and tryptophan UGC do not)
- "Stop" is encoded by UGA, UAG, UAA: no amino acid or tRNA.
- Short molecules, 70-90 nts long.
- All terminate with CCAOH-3' to which an amino acid can be covalently attached.
- Contain unusual nucleotides, which are modifications of the purine/pyrimidine bases or ribose
sugar, such as methylations, reductions, altered site of sugar linking to base.
Examples include:- thymidine (uridine with C5 methyl)
- methylated guanosines, methylated adenosines
- inosine and methylinosine (modified purines)
- pseudouridine (ribose sugar attached to uridine in the C-5 rather than C-1 position)
- dihydrouridine (uridine reduced at the C5-C6 double bond).
- Function of modifications
- control specific folding of the tRNAs. Some modifications are universal, and are therefore found in all tRNAs. These modifications contribute to the secondary structure (cloverleaf) shape of tRNAs, and the tertiary (L-shape) structure as well. Other modifications are specific to members of a family of tRNAs, and define them as such to the molecular machinery that covalently attaches a specific amino acid. Family specific modifications often serve as recognition signals for aminoacyl tRNA synthetases.
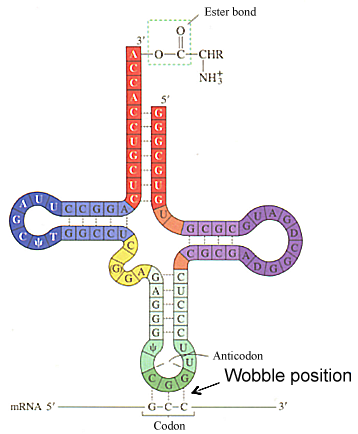
All tRNAs possess a common secondary and tertiary stem-loop structure that is critical for their function. A typical tRNA has the following secondary structure: a T-pseudouridine-C-G loop (TψCG loop - darker blue), a dihydrouracil or D-loop (darker purple), and an anticodon loop darker green). The anticodon loop contains the three complementary nucleotides that basepair with a specific codon in the mRNA (darkest green). A given tRNA interacts with different codons that specify a given amino acid due to nonstandard or wobble basepairing in the 3rd position of the codon with the 1st position of the anticodon.
- Genetic code is almost universal:
- same for prokaryotes and eukaryotes
- in mitochondria, the codon AUA encodes methionine rather than isoleucine, and AGA/G signals stop rather than arginine.
DECODING OF mRNA
- The genetic code is read during translation by tRNAs that have 3-base anticodons complementary to codons in mRNA.
- "Wobble" during reading of the mRNA allows some tRNAs to read multiple codons that differ only in the 3rd base.
- Mammalian cells produce more that 150 tRNAs
- Because the genetic code is degenerate, there often must be flexibility or "wobble" in the base pairing at the third position of the codon
- 3' end with respect to the mRNA
- 5' end with respect to the tRNA
- Degeneracy can be determined in two ways:
- there can be several tRNA for each amino acid
— each has a specific anticodon loop sequence - inosine (I) can act as a "wild card"
— can decode multiple codons
— I can base-pair with A, U, and C
- there can be several tRNA for each amino acid
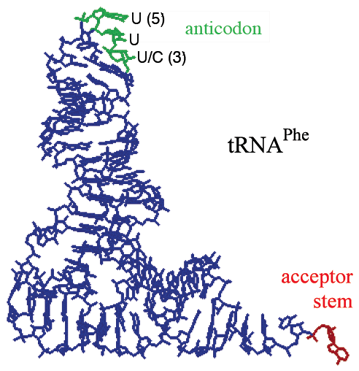
tRNA Tertiary Structure
- Identical position and type of unusual nucleotides in each tRNA family: defiines the tRNA family
- The specific tRNA structure and modified nucleotides are recognized by an enzyme (aminoacyl synthetase)
- There is one aa synthetase per amino acid/tRNA family
tRNAs Contain Modified Bases
— types and positions determine tRNA families 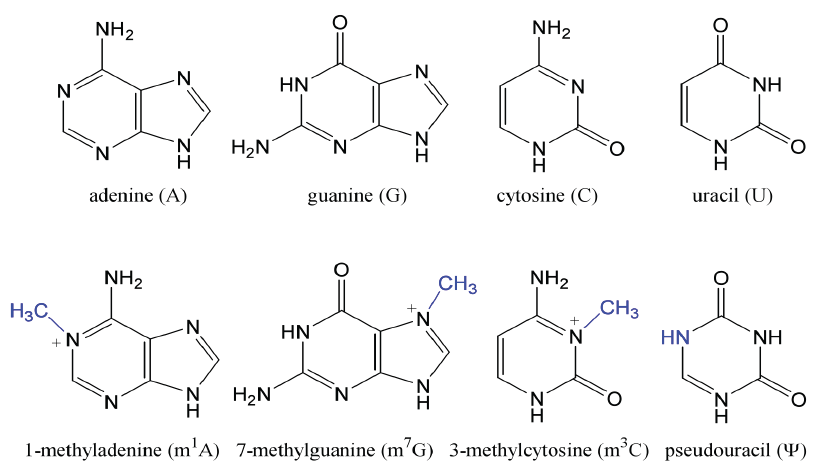
Aminoacyl-tRNA Synthetases Couple Amino Acids To tRNAs
Synthetase facts:
- Specific tRNAs are "charged" with specific amino acids by aminoacyl—tRNA synthetases - one synthetase for each tRNA family or type (eg. 1 synthetase for all of 6 of the Leu tRNAs)
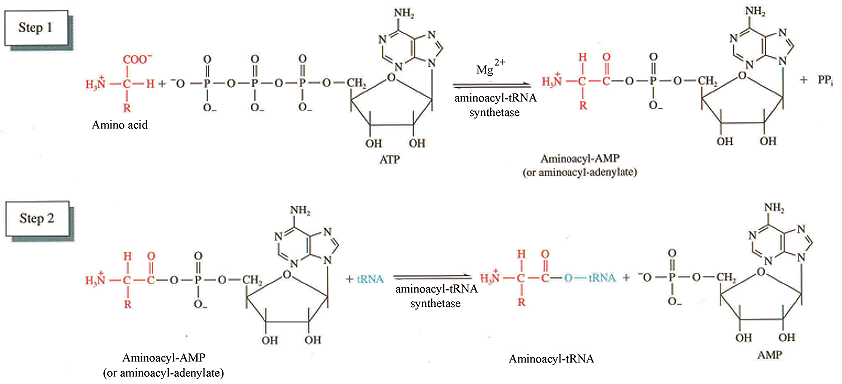
- Aminoacyl tRNA synthetases catalyze the reactions:
amino acid + ATP —> aminoacyl-AMP + PPi
aminoacyl-AMP + tRNA —> AMP + aminoacyl-tRNA
- Attachment of the amino acid is to the 3’OH of the A residue ribose sugar in the conserved CCA sequence on tRNA (acceptor stem).
- Energy in this bond is utilized later for polypeptide synthesis.
- Synthetases recognize different characteristics of tRNAs: unusual bases and anticodon, tertiary structure.
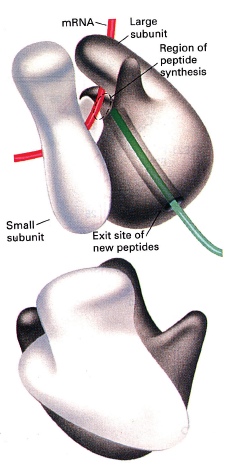
The active site (region of peptide synthesis) of the ribosome is at the interface of the small and large subunits
The active translating (elongating) ribosome is like a bead on a string
PROKARYOTIC AND EUKARYOTIC RIBOSOMES
Ribosomes are complicated structures consisting of ribosomal RNAs and proteins that associate into a precise structure with multiple enzymatic activities. The ribosomes of prokaryotes, eukaryotes and organelles (such as mitochondria) all perform the same function and are structurally quite similar. In evolution, ribosomes from prokaryotes and eukaryotes are unrelated at the protein level, but are highly related at the rRNA level.General Features in Common Between Eukaryotic and Prokaryotic Ribosomes
- 2 ribosome subunits, a small and large subunit.
- Consist of protein and RNA only.
- Ribosomal RNAs (rRNAs) are highly related between prokaryotes and eukaryotes, whereas ribosomal proteins (r-proteins) are not.
- Enzymatic functions of ribosomes involved in peptide synthesis are associated with rRNAs rather than r-proteins. r-proteins are thought to fine-tune and enhance function of rRNAs under physiological conditions.
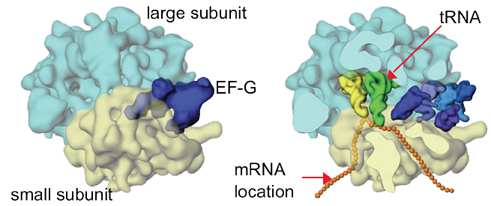
A tRNA (orange) is shown base pairing with part of mRNA (gold) on left and extending into the ribosome's peptidyltransferase center on right. Click To View Larger
The Functions Of Ribosomes In Translation Are Primarily Associated With rRNAs Rather Than rProteins.
The rRNAs: The Ribosome Has Three tRNA Binding Sites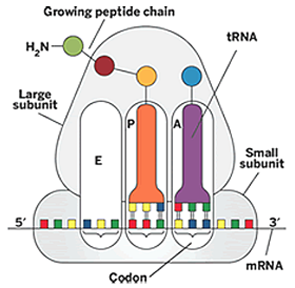
A site – amino acyl site, contains new incoming AA-tRNA
P site – peptidyl site, contains growing peptide chain
E site – exit site, site at which AA discharged tRNA is removed
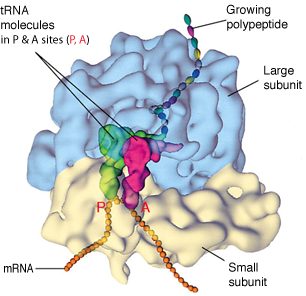
Space-Filling Model Of Translating Ribosome
During protein synthesis, incoming tRNA (purple) carrying the next amino acid (blue sphere) enters the A site if its anticodon is complementary in sequence to the codon on mRNA.
The reaction (not shown) between A-site tRNA and P-site tRNA (orange) extends the peptide chain by one amino acid unit.
The amine from a new amino acid on the tRNA bound at the A site attacks a carbonyl at the end of the growing peptide chain, which is attached to the tRNA bound at the P site.
Some Antibiotics Block Ribosome FunctionSpecific segments of 16S & 23S rRNAs have been identified that correspond to the A and P sites.
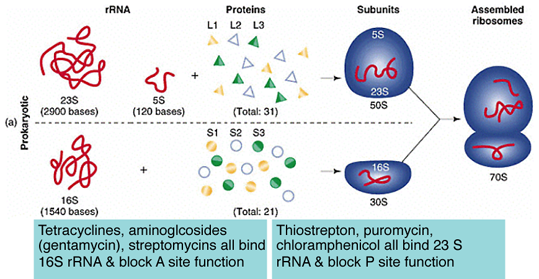 Many antibiotics act by binding or blocking rRNA activity within these enzymatic sites.
Many antibiotics act by binding or blocking rRNA activity within these enzymatic sites.
- Thiostrepton — binds 23S rRNA (residue A1067) and prevents 50S subunit association. Methylation of A1067 provides resistance.
- Puromycin (aminoacyl-tRNA analogue) — blocks domain V of 23S rRNA responsible for peptidyl transferase activity; blocks peptide bond formation.
- Tetracycline — probably binds 16S rRNA at A892, same site that tRNA binds in the A-site.
- Streptomycin — probably binds and blocks activity of 16S rRNA near nt 900. Activity is similar to tetracycline.
- Aminoglycosides (neomycin, gentamicin, kanamycin, hygromycin) — bind specific sites in the A-site contributed by 16S rRNA, prevents translocation of the ribosome along the mRNA. Resistance is associated with mutation of sites in this region.
- Edeine — binds P-site within 16S rRNA, prevents tRNA association with 30S subunit.
- Chloramphenicol & carbomycin — bind domain V loop in 23S rRNA, inhibit peptidyl transferase activity. Resistance is associated with mutation in this region.
MECHANISM OF PROTEIN SYNTHESIS — OVERVIEW
Protein synthesis can be divided into 6 stages:- Amino acid activation: tRNA is charged by covalently linking it to its cognate amino acid (above).
- Formation of initiation complexes: association of mRNA, ribosomal subunits and initiation factors.
- Initiation of translation: assembly of stable ribosome complex at the initiation codon.
- Chain elongation: polypeptide synthesis by repetitive addition of amino acids to the nascent (growing) chain.
- Chain termination: release of nascent polypeptide.
- Ribosome dissociation: subunits separate before initiating new round of translation.
INITIATION COMPLEX FORMATION
Initiating tRNA
Facts:
- Translation generally initiates with a Met encoded by AUG (prokaryotes & eukaryotes).
- Special initiating tRNA carries Met to AUG codon.
- In bacteria the initiating Met is modified, while attached to the tRNA, to contain an N-formyl group. It is referred to as N-formylMet (tRNAfmet).
- The formyl group blocks acceptance of a growing peptide chain.
- Elongating met-tRNA is distinct (tRNAmet), and the Met is not modified. The formyl group is always removed from bacterial proteins.
- In eukaryotes the initiating Met is not modified (tRNAimet).
- The initiating Met is removed from roughly half of bacterial proteins, and from some eukaryotic proteins
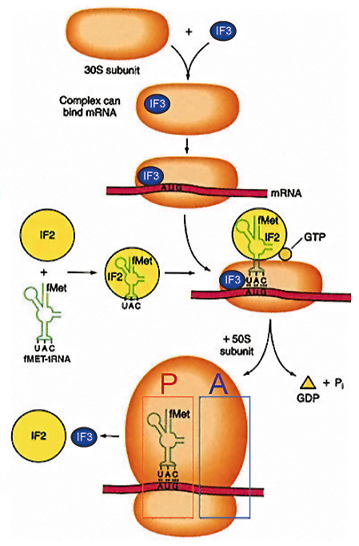
Mechanism Of Translation Initiation
Anti-association factors IF1 and IF3 bind the 30S subunit and prevent 50S subunit association. Eukaryotic initiation factors eIF1 and eIF3 are similar and they have the same functions. 30S subunit associates with tRNAfmet, GTP and IF2 to form a ternary complex. Association of ternary complex components, 30S ribosome and mRNA in prokaryotes takes place in any order. In eukaryotes it is highly ordered (as described later).
Initiation requires the assembly of the mRNA on the ribosome at the initiating AUG for translation
- Small subunit first associates with mRNA
- A charged Met tRNA associates with the small subunit in combination with initiation factor IF2 and GTP (ternary complex)
- The large subunit is recruited to finish the complex: first tRNA is present in the P site of the ribosome
INITIATION OF TRANSLATION
- Joining of small and large ribosomal subunits with the mRNA creates a 70S ribosome initiation complex.
- Initiation is guided by nucleotide pairing between a sequence in the mRNA and the 3’
end of 16S rRNA, in a process called the Shine-Dalgarno (S/D) interaction:
mRNA5’---------UAAGGAGG(5-10 nts)AUG------ 16S rRNA 3’(OH)--------AUUCCUCC----------
- The level of translation of the mRNA is controlled by the S/D interaction, which is the
extent of complementarity between mRNA:rRNA, and the most optimal AUG position
(5-10 nts downstream of the S/D element). This explains why prokaryotic ribosomes can
initiate protein synthesis internally. As a consequence, prokaryotic mRNAs are generally
multicistronic, encoding more than one polypeptide. This also explains why the ternary
complex can form on ribosomes after the subunits associate in prokaryotes, because there
is no need for tRNAfmet to provide anticodon identification of the initiating AUG.
- Greater than 90% of initiation codons are AUG
- Initiating codons other than AUG: infrequently GTG ("Valine") and TTG ("Leucine")
- A methionine is still incorporated as the first amino acid
- S/D sequence controls initiation in two ways: spacing of the AUG fron the S/D (5-10 nucleotides); complementarity of interaction with the 3' end of the 16s rRNA
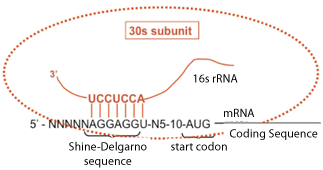
Base-Pairing of Shine/Delgarno Sequence With Its Complementary Sequence Near The 3' End Of 16s Ribosomal RNA
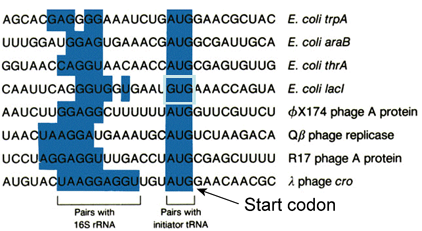
Examples Of Shine/Delgarno Sequences In Selected mRNAs
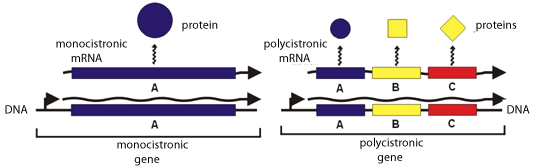
The Shine-Delgarno Interaction Permits The Development Of Operons — Genetic Grouping Of Genes Of Related Function
A single mRNA can encode multiple proteins — polycistronic mRNA — of related function, and their translation can be temporally orchestrated when needed.Eukaryotes initiate translation quite differently. In eukaryotes there is no such sequence or S/D interaction (at least routinely). In fact, the Shine Dalgarno sequence is specifically missing from the 3’ end of eukaryotic 18S rRNA.
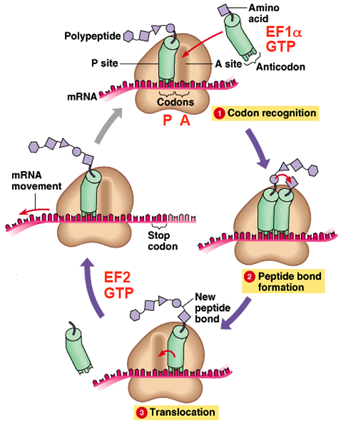
Steps In Translation Elongation
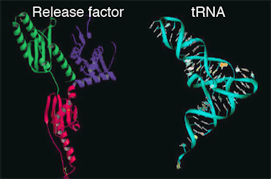
Release factor has a 3-dimentional shape similar to that of tRNA
BASIC MECHANISM OF TRANSLATION ELONGATION
- The ribosome selects aminoacylated tRNA (
 codon recognition).
codon recognition). - EF1α and GTP are bound to aminoacylated tRNA
- The ribosome catalyzes formation of a peptide bond (
 peptide bond formation).
peptide bond formation). - Translocation is dependent on EF2 and GTP hydrolysis
( translocation).
translocation). - Many ribosomes may translate mRNAs simultaneously on the same mRNA molecule.
MECHANISM OF TRANSLATION TERMINATION AND RIBOSOME DISSOCIATION |
||
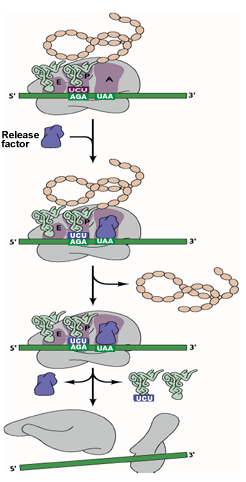 |
Translation is terminated at one of three stop codons (UAA, UAG & UGA). | |
| The termination codon at the A site is recognized by the release factor instead of a tRNA. | ||
| The release factor binds the termination codon. | ||
| The peptide chain is then released followed by dissociation of the tRNA and the ribosome. | ||