James' Unwanted Blood Clots
The laboratory report states that James has half the normal activity of antithrombin III.
The following pages illustrate how thrombin works and how antithrombin III inhibits it.
Thrombin is a typical member of the serine protease class of enzymes.
Serine proteases have:
- a catalytic site. The catalytic site has the same structure in all serine proteases.
- a substrate recognition site. Different serine proteases have different substrate recognition sites.
- thrombin has other sites that are not covered in this tutorial. Some of these sites interact with small molecules and other proteins that modify thrombin activity while others interact with cellular receptors.
Thrombin's catalytic site
Prothrombin is cleaved to produce two chains that interact to form thrombin: a light chain and a heavy chain.
Thrombin's catalytic properties reside in the heavy chain.
For clarity, only the heavy chain is shown here and in subsequent structures in this tutorial.
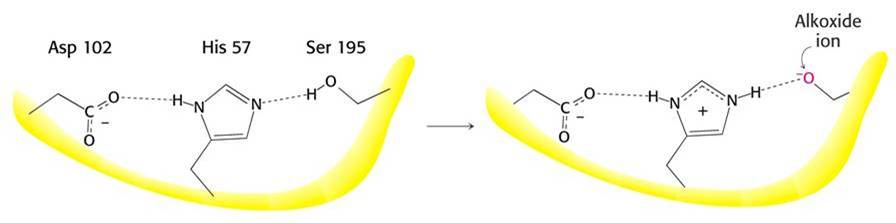
Like all serine proteases, thrombin's catalytic site contains a serine, a histidine and an aspartic acid.
These three residues function together as a "catalytic triad".
Thrombin's tertiary structure brings these three residues together precisely at the substrate binding site.
They are positioned so that they work together to catalyze the hydrolysis reaction that cleaves the peptide bond of the substrate.
|
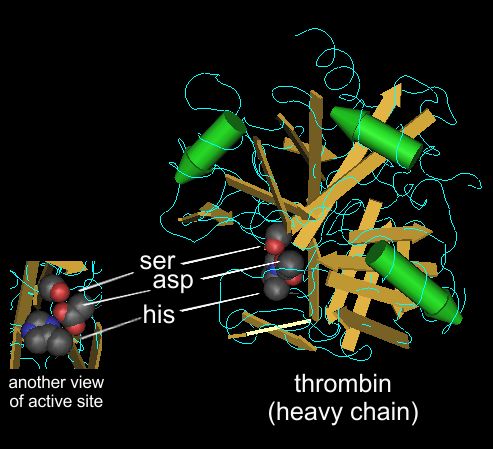
This is a molecular structure of thrombin as determined by x-ray crystallography.
The gold arrows and green cylinders show elements of of secondary structure (amino acids in sheets are gold and amino acids in alpha-helices are green)
This structure (or any other protein structrue that has been determined) can be viewed as a 3-D model that you can rotate and manipulate on your computer screen.
These protein structures are available to the public via the "structures" section of the NCBI databases (NCBI = National Center for Biotechnology Information).
The NCBI databases also include "pubmed" and "OMIM".
This is structure 3BV9, which can be accessed here
|
You do not need to memorize the serine protease reaction mechanism, but seeing it may help your understanding of how the exact positioning of amino acids within an enzyme can create a catalytic site.
The mechanism, as shown in detail in Dr. D'Eustachio's lectures, depends on the formation of a reactive environment created by the side chains of a serine, histidine and aspartic acid.