INTRODUCTION
DNA replication is the process by which the DNA double helix is separated and then copied by specialized enzymes to give two identical daughter molecules. Proper control over the initiation, elongation, and termination of DNA replication is essential because life requires the genetic information encoded in DNA to be stable. Having detailed understanding of DNA replication is important because it is a fundamental biological process. It is also necessary to understand DNA replication for the following reasons:
- Defects in replication can lead to numerous diseases, including many forms of cancer.
- Specific limitations with the replication machinery give rise to certain classes of mutations such as the trinucleotide repeat expansion disorders (e.g., fragile X syndrome).
- DNA replication in cells experiencing unregulated growth (e.g., cancer cells) is often a target of anti-cancer agents.
- Replicative processes are involved with the repair of DNA damage due to radiation, mutagens, etc, that when faulty can cause disease (e.g., Xeroderma pigmentosum).
- The enzymes involved with DNA replication are among the most powerful tools in use for genetic engineering and for clinical diagnosis (e.g., PCR).
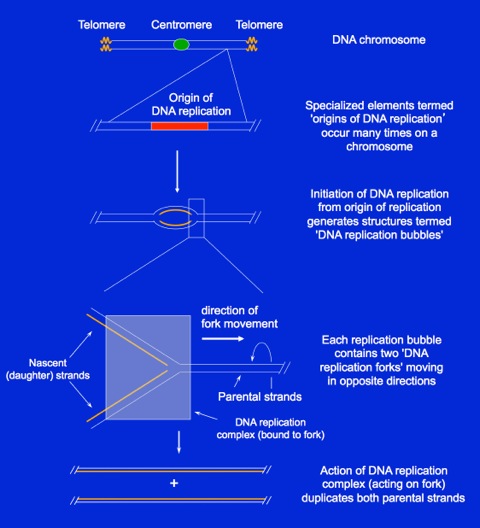 Overview Of DNA Replication
Overview Of DNA ReplicationOVERVIEW OF DNA REPLICATION
The figure at the right shows a typical eukaryotic chromosome. On each end of the chromosome are specialized structures termed telomeres. Each chromosome also contains a centromere, a DNA element that is involved in separating the two sister chromosomes during mitosis. In human cells, the centromere is generally located towards the center of each chromosome. Each chromosome also contains many ‘origins of DNA replication’ that support the initiation of DNA replication. The activation, or “firing”, of a replication origin leads to the formation of a DNA replication bubble that contains two replication forks moving in opposite directions. Elements of a DNA replication fork are conserved in all systems. DNA replication at the forks occurs in organized complexes that move along the DNA, opening the DNA at the front and synthesizing a nascent strand on each parental strand.OKAZAKI FRAGMENT SYNTHESIS
DNA synthesis always occurs in the 5’ to 3’ direction. Because of this, the DNA replication fork has an asymmetric structure. Only one daughter strand can be synthesized continuously, and this is termed the leading strand. The direction of the DNA polymerization reaction is the same as the direction of DNA fork movement. Therefore, the nascent leading strands can be synthesized in very long tracts containing many thousands of base pairs. The synthesis of a leading strand can continue until 1) the replication machinery meets a fork moving in the opposite direction (i.e., from another replication origin), 2) the fork completes DNA synthesis at telomeres at chromosome ends, or 3) the fork encounters damaged DNA that stalls fork progression.
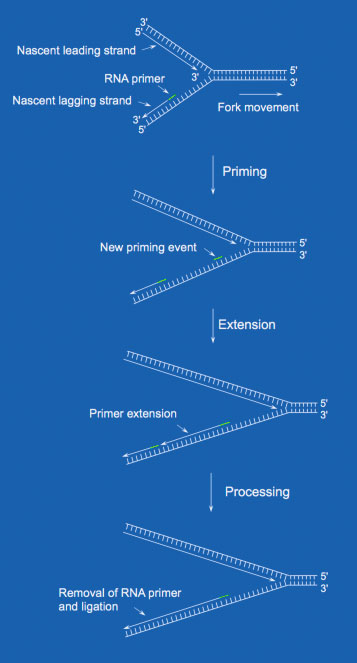 Synthesis of Okazaki fragments
Synthesis of Okazaki fragmentsEach Okazaki fragment length in bacteria (i.e., E. coli) is about 1000 nts in length on average, while in humans they have an average length of ~200 nts in length. Regarding replication rates, bacteria incorporate 500 nts per strand per second. In eukaryotes, only 50 nts are incorporated per strand per second. Even though humans have 1000 times as much DNA per cell as a bacterial cell, the slower replication rate in eukaryotes is thought to reflect the need for additional time to prevent the occurrence of errors.
THE DNA POLYMERASE ENZYME
DNA polymerases are those enzymes that incorporate the DNA precursors, deoxyribonucleoside triphosphates (dNTPs - dATP, dGTP, dCTP, and dTTP), into a nascent strand, using hydrogen-bonding interactions with nucleotide bases on the template strand.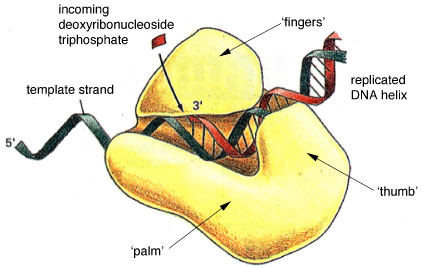 The structure of DNA polymerase resembles a human right hand
The structure of DNA polymerase resembles a human right handThe structure of various DNA polymerases have been solved by X-ray crystallography. These structures indicate that the “typical” DNA polymerase resembles a human right hand, in which the thumb, palm, and fingers clasp the DNA. In this way, the DNA is properly positioned for association with the polymerase active site, and catalytic efficiency is promoted.
NUCLEOTIDE INCORPORATION BY DNA POLYMERASES
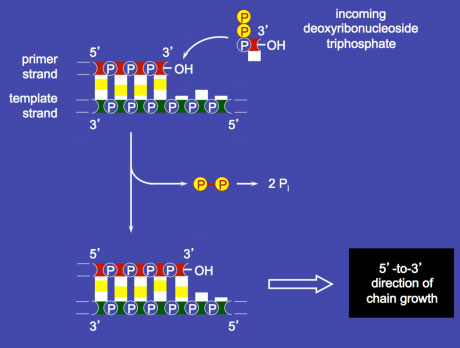 Nucleotide incorporation by DNA polymerase
Nucleotide incorporation by DNA polymerase
TAUTOMERIZATION
The DNA polymerase has a property (termed polymerase specificity) that allows the enzyme to detect an incorrectly paired dNTP before it is incorporated into the growing chain. Even so, each of the nucleotide bases can exist in other isomeric forms. The nucleotide bases in DNA can spontaneously undergo a transient rearrangement of bonding, termed a tautomeric shift, to form a structural isomer (or tautomer) of the base. The tautomerization of bases is a relatively rapid equilibrium reaction, but greatly favors the common form.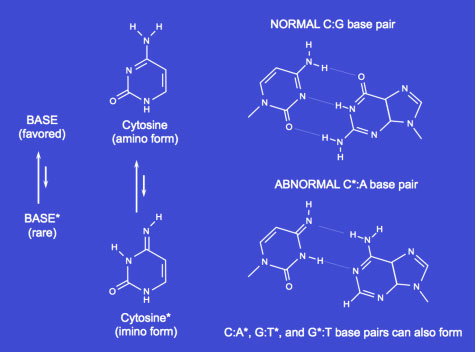 Tautomerization of nucleotide bases results in aberrant base pairing
Tautomerization of nucleotide bases results in aberrant base pairing
DNA POLYMERASE PROOFREADING
An important cellular process required for high fidelity DNA replication is DNA polymerase proofreading. This mechanism exists to “edit&rdsquo; mistakes in nucleotide incorporation. Along with the 5' to 3' catalytic subunit for DNA synthesis, DNA polymerases that support proofreading also contain a 3' to 5' exonuclease. This latter activity allows removal of single nucleotides from the 3' end of a DNA strand as follows.
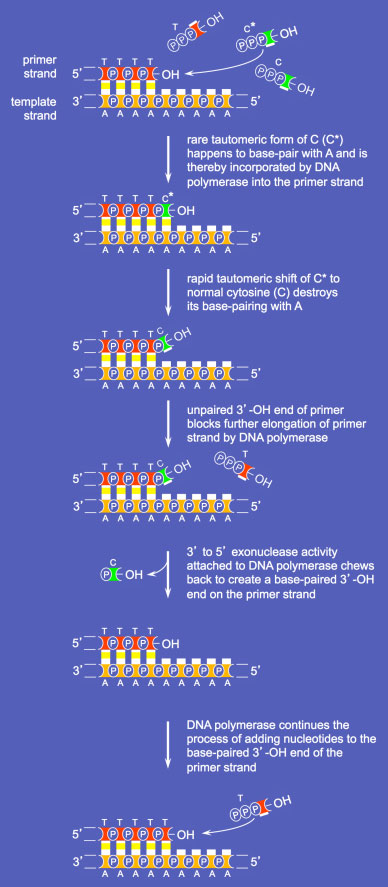 Proofreading by the DNA Polymerase 3' to 5'
Proofreading by the DNA Polymerase 3' to 5'exonuclease activity
The major DNA polymerases acting during DNA replication all have proofreading activities. These and other DNA polymerases use magnesium at their active catalytic sites for DNA synthesis. Of note, higher than normal concentrations of other divalent metals such as Mn2+, Co2+, Cd2+, Be2+ can displace Mg2+ from the active site, reducing the replicative fidelity of the polymerase. The Mg2+-displacing metals have all been implicated as carcinogens. For example, workers who are exposed to high levels of cadmium are subject to a higher than normal rate of prostate cancer, because polymerase misincorporation occurs at a higher than normal rate. The proofreading activity of the DNA polymerase is unable to effectively repair the greater number of polymerase misincorporations.
HUMAN DNA-DIRECTED DNA POLYMERASES
There are a number of different DNA polymerases in mammalian cells. A table of the 15 DNA polymerases in human cells is shown (from Lange et al. 2011. Nat. Rev. Cancer 11:96-110). A major point of the table is that human cells encode multiple DNA polymerases. The most important human DNA polymerases for general DNA replication are: DNA polymerase α (which has an associated DNA primase activity, needed to make an RNA primer for Okazaki fragment synthesis); DNA polymerase δ which synthesizes the bulk of the Okazaki fragment on the lagging strand; and DNA polymerase ε which synthesizes the leading strand.The other DNA polymerases have other specialized functions. For example, DNA polymerase γ localizes to mitochondria, where it is involved in the replication and repair of mitochondrial DNA. As a second example, DNA polymerase β is a protein involved in so-called base excision repair, in which only one or two nucleotides are synthesized. Most of the other DNA polymerases have been identified more recently, and the characterization of their functional role is ongoing.
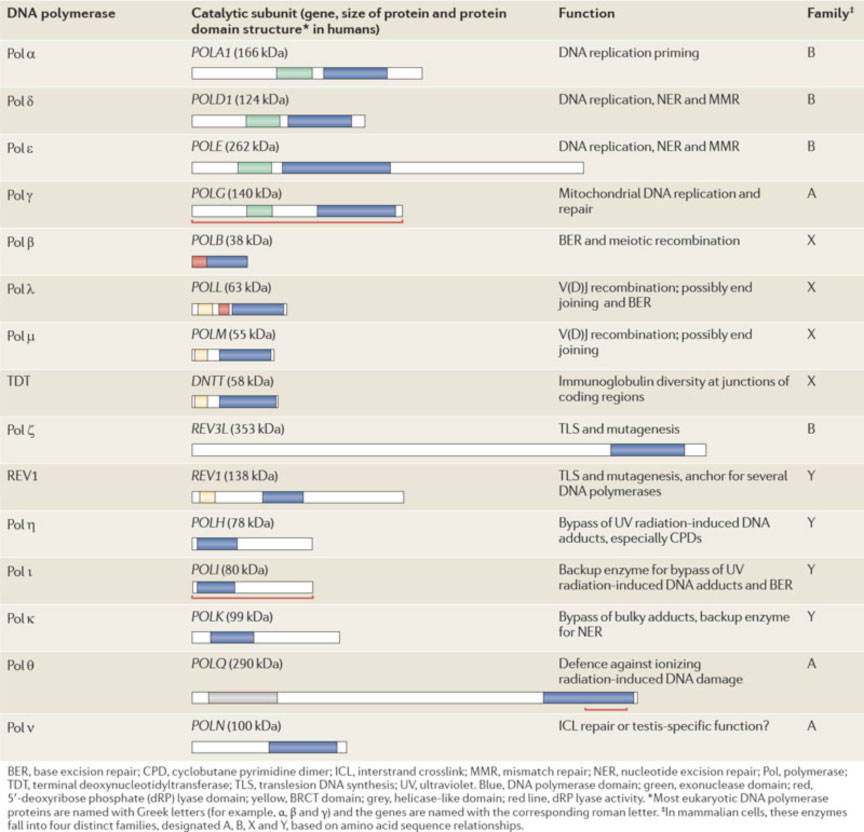
IMPORTANT DNA REPLICATION FACTORS
In addition to the DNA polymerases that incorporate dNTPs into the growing DNA chain, a number of other proteins are required to increase DNA polymerase activity, either directly or indirectly during DNA replication.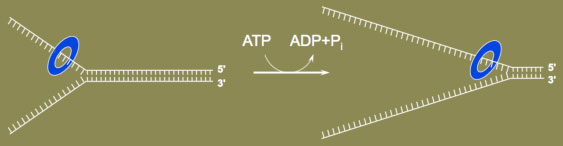 DNA helicases unwind the DNA double strands
DNA helicases unwind the DNA double strandsOne important class of factors is DNA helicases. DNA helicases are protein motors that use the energy of ATP hydrolysis to unwind DNA, for example, at the DNA replication fork (see figure; blue ring encircling one DNA strand is the DNA helicase). Many types of DNA helicases exist that act in additional capacities at the replication fork, or that function in DNA repair or DNA recombination. Importantly, mutation of human genes encoding DNA helicases can cause disease. For example, Bloom's syndrome was initially identified in the 1950s by NYU dermatologist David Bloom. In the 1990s, the BLM gene that when mutated gives rise to the syndrome was identified and found to encode a DNA helicase. An important clinical consequence caused by mutation of human BLM is genomic instability, and a greatly increased incidence of cancer.
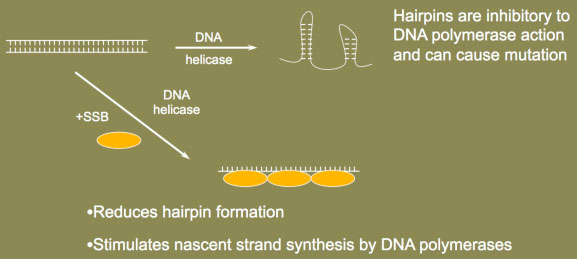 Single-Stranded DNA-Binding Protein prevents hairpin formation
Single-Stranded DNA-Binding Protein prevents hairpin formationA second essential factor at the replication fork is the single-stranded DNA-binding protein, or SSB. As the name suggests, these proteins bind to single-stranded DNA (ssDNA) with high affinity after the ssDNA is formed, for example, by a DNA helicase. SSBs are needed for DNA replication and repair because ssDNA can fold upon itself to form structures such as DNA hairpins. During DNA replication, hairpin production would be most prevalent on the lagging strand because ssDNA would form prior to the synthesis of each Okazaki fragment. Hairpin formation is greatly inhibitory to DNA replication because they stall the movement of the DNA polymerases. The formation of such secondary structures on ssDNA can also cause an increase in DNA polymerase errors during DNA synthesis. As evidence of this, mutation of the mammalian SSB, a protein called RPA, causes defective DNA double-strand break repair, chromosomal instability and cancer in mice.
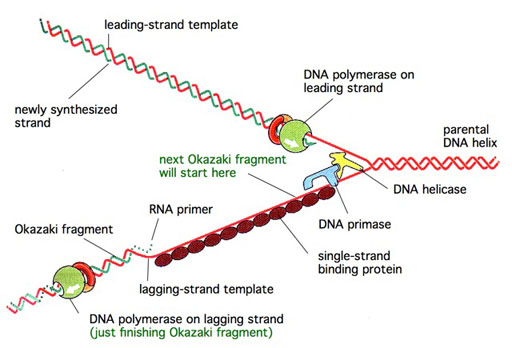 Proteins at the DNA replication fork
Proteins at the DNA replication forkTHE PROTEINS AT A GENERIC DNA REPLICATION FORK
A simple model showing the disposition of the major protein factors at a generic DNA replication fork is shown (see Figure). A DNA helicase acts at the front edge of the fork to unwind the DNA. The DNA polymerases are present on both the leading and lagging strands, synthesizing the nascent DNA strands. An enzyme called DNA primase serves to initiate each Okazaki fragment on the lagging strand. A number of SSB molecules would also be bound to the exposed ssDNA on the lagging strand template, with the SSBs acting to stimulate DNA synthesis, and to reduce errors by the DNA polymerases.GENERATION OF TORSIONAL STRESS
The replication fork proceeds relatively quickly along the DNA, moving at a rate of 500 nucleotides per second in bacteria, and 50 nucleotides per second in human cells. Because there are 10 base pairs per helical turn of DNA, unwinding of the DNA double helix would occur at a rate of 50 rev/sec in bacteria, and 5 rev/sec in humans. This rapid DNA unwinding generates torsional stress ahead of the fork, because the covalent nature of each DNA strand prevents easy rotation of one strand with respect to the other. As the torsional stress builds in the DNA, the replication fork would have a difficult time moving along the DNA because more work is needed to unwind the DNA.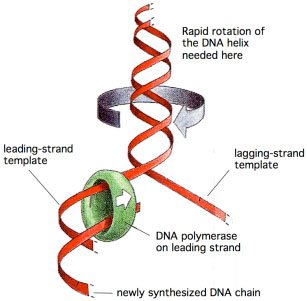 DNA ahead of the fork rotates at a rate
DNA ahead of the fork rotates at a rate of 50 rev/sec, creating torsional stress
MECHANISM OF TYPE I TOPOISOMERASES
Type I topoisomerases reversibly nick the DNA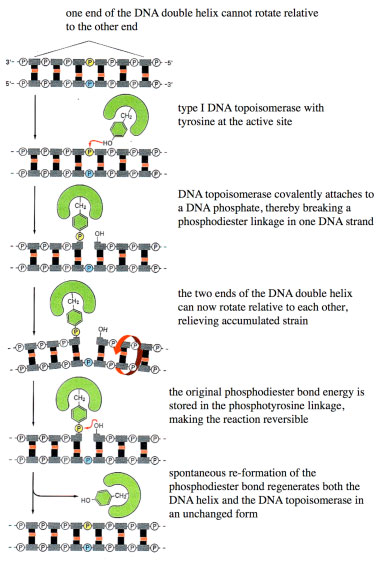 Type I Topoisomerases releive torsional stress
Type I Topoisomerases releive torsional stressNEED FOR TYPE II TOPOISOMERASES DURING MITOSIS
The process of eukaryotic DNA replication results in two interlinked daughter chromosomes (see Figure). When the daughter chromosome pairs are being segregated to opposite poles during mitosis, the interlinks must be quickly removed to allow the two chromosomes to separate. Type II topoisomerase are enzymes that catalyze removal of the interlinks, using a mechanism involving the passage of one DNA helix through another. In the absence of topoisomerase II, cytokinesis cannot properly occur, causing the chromosomes to fragment into shorter pieces. This fragmentation is termed mitotic catastrophe.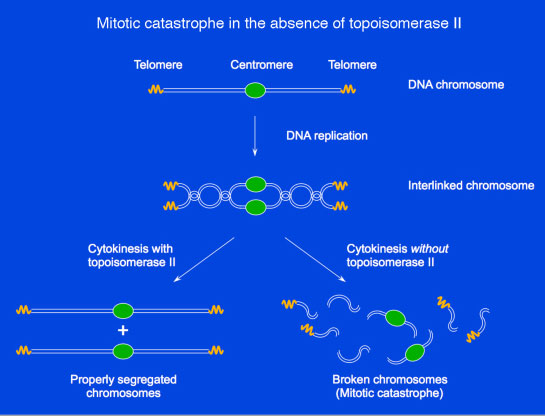 Type II topoisomerases remove interlinks resulting from DNA replication
Type II topoisomerases remove interlinks resulting from DNA replicationVarious therapeutic agents are directed against both type I and type II topoisomerases, because of their critical roles during DNA replication and mitosis. Examples of such agents will be discussed during lecture.
INITIATION OF EUKARYOTIC DNA REPLICATION
Eukaryotic DNA replication is initiated by the binding of ORC (origin recognition complex) at a replication origin. With the aid of other initiation factors (i.e., the Cdc6 and Cdt1 proteins) the MCM complex is delivered to the origin, generating the pre-RC (pre-replicative complex). After the action of two kinases (Cdk and Cdc7) that phosphorylate pre-RC components, additional replication factors are recruited to the complex. Using a mechanism still being elucidated, the replication machinery unwinds the bound double-stranded DNA. The DNA polymerases then associate with the two nascent forks to begin DNA synthesis, with each fork moving away from the origin.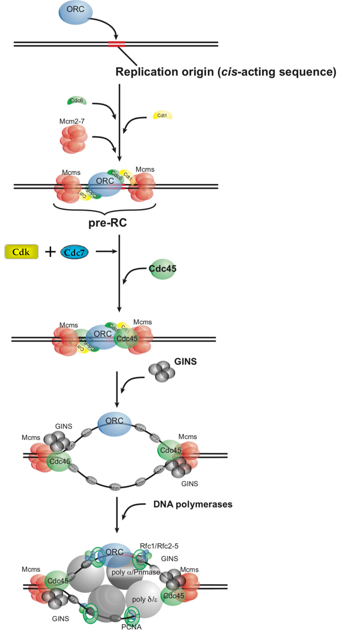 Mechanism of DNA Replication
Mechanism of DNA ReplicationTELOMERES AND TELOMERASE
The ends of eukaryotic chromosomes contain specialized protein-DNA structures called telomeres. At the DNA level, telomeres normally contain large numbers of short repeated sequences. In vertebrate cells, the telomeric repeat sequence is TTAGGG (5' to 3' toward the chromosome end). The total length of telomeres in humans depends on cell type and age, and generally ranges from 5 to 15 kb. Telomere repeats are synthesized by a specialized enzyme termed telomerase, which is an RNA-dependent DNA polymerase. The RNA template to allow extension of the repeats is a component of the telomerase complex.Normal human somatic cells express very low levels of telomerase. As a person ages, the telomere shortens. In contrast, germline cells and many tumor cells express telomerase and thereby maintain telomere length. Transformation of normal somatic cells with oncogenes eventually leads to a crisis, because the increased number of cell division cycles causes telomeres to become critically short. Although most cells die at this point, the rare cell that expresses telomerase activity survives. Although upregulation of telomerase activity is routinely seen in cancer cells, other mechanisms have also been found to maintain telomeres, a critical step in cell immortalization.
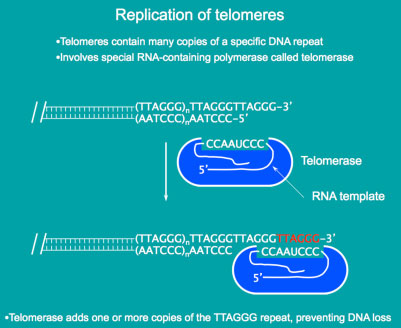 Telomeres and Telomerase - maintaing DNA at the ends
Telomeres and Telomerase - maintaing DNA at the endsof chromosomes
OVERVIEW OF DNA REPAIR
Mechanisms exist to repair damaged DNA, and to correct mispaired DNA. In most cases, DNA repair causes the replacement of portions of one of the two DNA strands. The length of the DNA replaced can range from a single nucleotide to over 1 kilobase of DNA.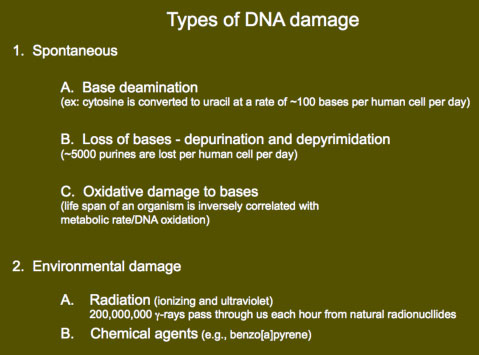 DNA damage
DNA damageDNA damage often does not result from external sources (e.g., radiation exposure) but rather occurs because of the chemistry of DNA. For example, in each human cell, 100 cytosine bases spontaneously deaminate per day to give uracil. Other types of deamination also happen including the conversion of adenine to hypoxanthine, and guanine to xanthine. Another major reaction is the loss of bases from the sugar-phosphate backbone, that is, depurination and depyrimidation. It has been determined that 5000 purine bases are lost daily from the DNA of each human cell because of hydrolysis of the N-glycosyl linkage between the base and deoxyribose moiety. There is also a significant amount of oxidative damage to bases in cells. Interestingly, there is a correlation between rate of DNA oxidation and the metabolic rate of organisms. Oxidative damage possibly affects aging, because the life span of an organism is inversely correlated with the metabolic rate, and thus the rate of oxidative damage.
Although much DNA damage occurs spontaneously, damage can also be caused by the environment. Humans are often exposed to ultraviolet rays from sunlight, giving cyclobutane pyrimidine dimers (including thymine dimers), and to ionizing radiation from cosmic rays. It has been estimated that 100,000 cosmic ray neutrons and 200,000,000 gamma rays pass through our bodies every hour. DNA damage can also come from chemicals in the environment such as benzo[a]pyrene, a mutagen and carcinogen found in cigarette smoke.
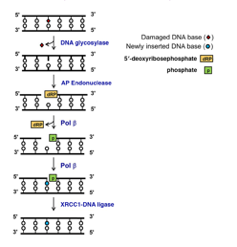 Short-Patch Base Excision Repair
Short-Patch Base Excision Repair
BASE EXCISION REPAIR
Base excision repair (BER) is an essential DNA repair pathway. This pathway is primarily responsible for removing small, non-helix-distorting base lesions from the genome. There are three sub-types of BER in humans but the most simple is Short-patch BER (see figure). The process begins in humans with a damage-specific DNA glycosylase – the human genome encodes 11 different enzymes – excising the damaged base. This generates an apurinic/apyrimidinic [AP] site that is incised by the AP endonuclease to create a single-strand break. The residual 5' deoxyribosephosphate (dRP) moiety is removed by DNA polymerase β (Pol β), which simultaneously fills the gap with a new nucleotide. The remaining nick in the phosphodiester backbone is sealed by a XRCC1-DNA ligase complex to complete short-patch BER. Human mutations in BER are occasionally found, for example, mutations in the DNA glycosylase MYH increase susceptibility to colon cancer. Along with repairing base lesions, BER also functions to remove thymine from G-T base pairs, which can form by spontaneous deamination of 5-methylcytosine in CpG residues. Incomplete repair of the G-T base pairs over an evolutionary time frame are thought to have caused the under-representation of CpG in the human genome, to less than one-quarter of the expected frequency.NUCLEOTIDE EXCISION REPAIR
Cyclobutane pyrimidine dimers such as thymine dimers are repaired by a process termed nucleotide excision repair (NER).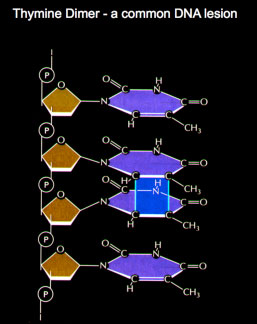 Thymine dimers are removed
Thymine dimers are removedby the Nucleotide Excission Repair
mechanism
MISMATCH REPAIR
The chief source of DNA alterations arising during normal DNA metabolism is the mispairing of bases during DNA synthesis, resulting in mismatches. Although DNA repair and recombination can lead to these changes,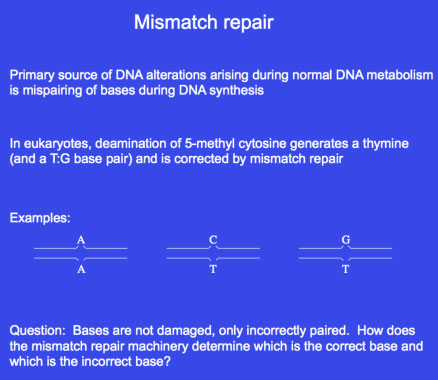 Repair of mismatched base
Repair of mismatched baseAn important point to note about mismatch repair is that the structure of the DNA bases is normal on both strands. Instead, there is a mismatch at one or more base that causes a bubble to form on DNA. Therefore, mismatch repair requires the mispair to first be recognized, and then the strand containing the incorrect nucleotide to be determined. In bacteria, identification of the nascent strand, necessarily containing the incorrect base, generally involves recognition of modification marks on DNA. How the new and old strands are distinguished in eukaryotes is not yet well understood.
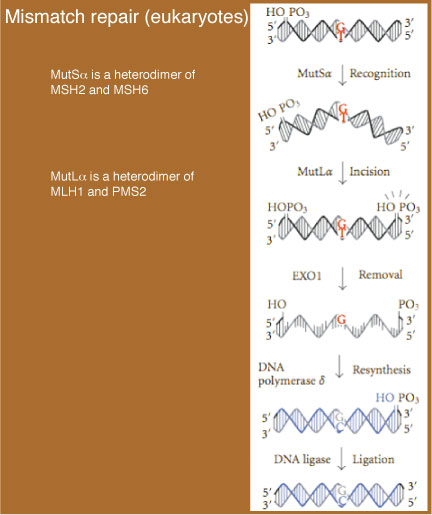 Mechanism of Eukaryotic Mismatch Repair
Mechanism of Eukaryotic Mismatch Repair
EUKARYOTIC MISMATCH REPAIR
Mismatch repair in eukaryotes is catalyzed by a number of factors.- In the first step, the mismatch is recognized by MutSα (a heterodimer of MSH2 and MSH6).
- After the repair machinery identifies the (nascent) strand containing the incorrect base, an incision (i.e., a nick) in that strand is made by MutLα (a heterodimer of MLH1 and PMS2). With this incision, removal of the mismatch can take place.
- An exonuclease (EXO1) then excises the DNA on the nascent strand between the nick and up to and including the mismatch. This excision can lead to the removal of over 1000 bp of DNA on that strand, generating a region that contains a ssDNA gap.
- In the final stage, DNA polymerase δ fills in the gap and,
- the remaining nick is sealed by DNA ligase.