DETERMINING THE AMINO ACID SEQUENCES OF PROTEINS
The three-dimensional shape and hence the functional properties of a protein are determined by its amino acid sequence. While we do not yet know how to predict these properties fully from a protein’s sequence, we can use sequence data to assign proteins to functional families, to identify specialized domains within proteins, and to determine likely effects of mutations that alter a protein’s sequence.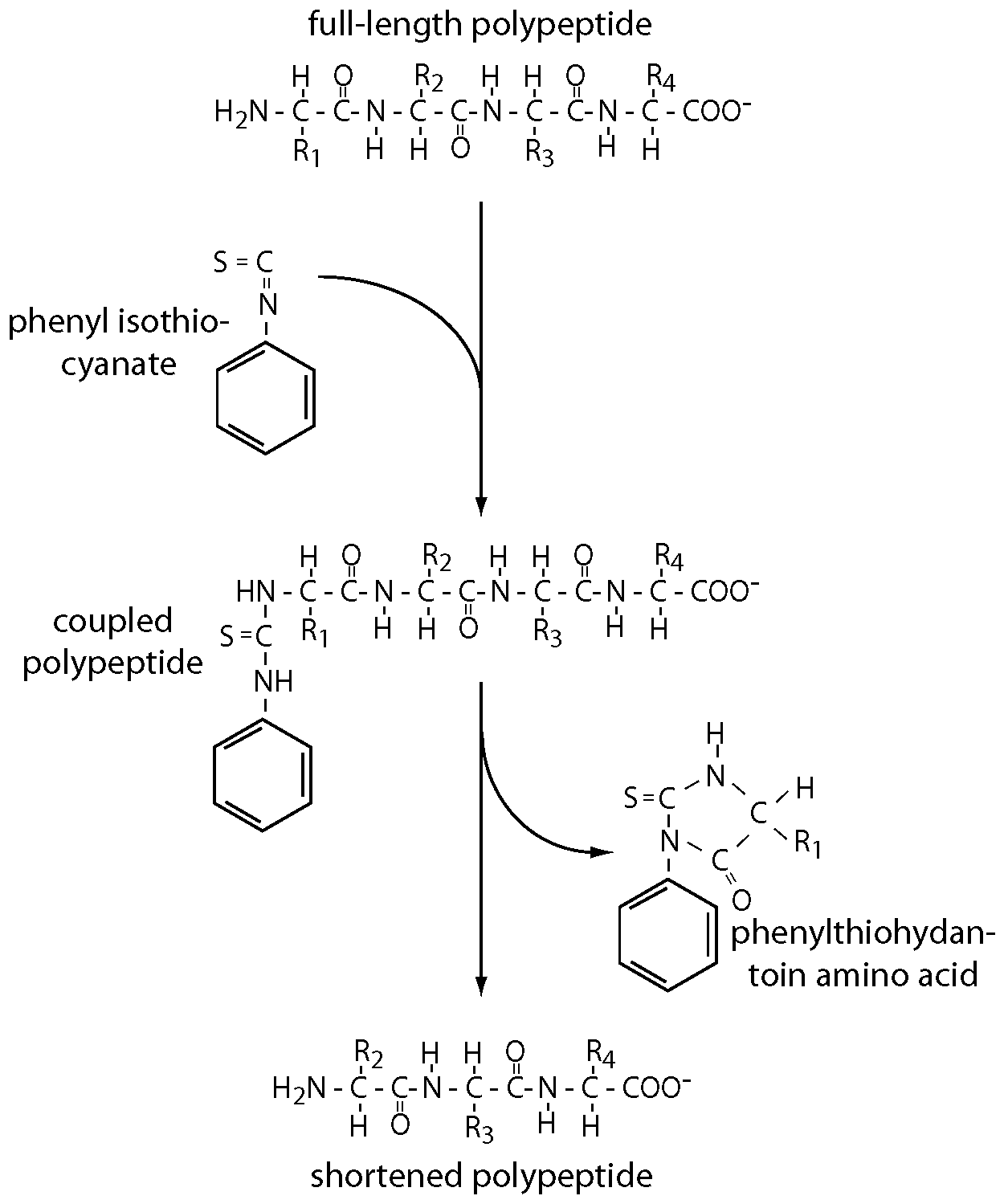
The chemical reactions are outlined in the diagram. Its key features are:
- Phenyl isothiocyanate couples to the free amino terminus of a polypeptide chain in a reaction that can be driven to completion.
- A second reaction that can also be driven to completion cleaves the coupled amino acid from the rest of the polypeptide as a phenylthiohydantoin derivative.
- This derivative can be separated from the polypeptide chain, which is now one residue shorter. Analytical techniques allow all 20 possible derivatives to be identified, and the remainder of the polypeptide meanwhile can be subjected to another round of coupling and cleavage.
- In this way, the sequence of a polypeptide can be read off, one residue at a time.